
- Presentation of images in cellprofiler analyst manual#
- Presentation of images in cellprofiler analyst software#
Presentation of images in cellprofiler analyst software#
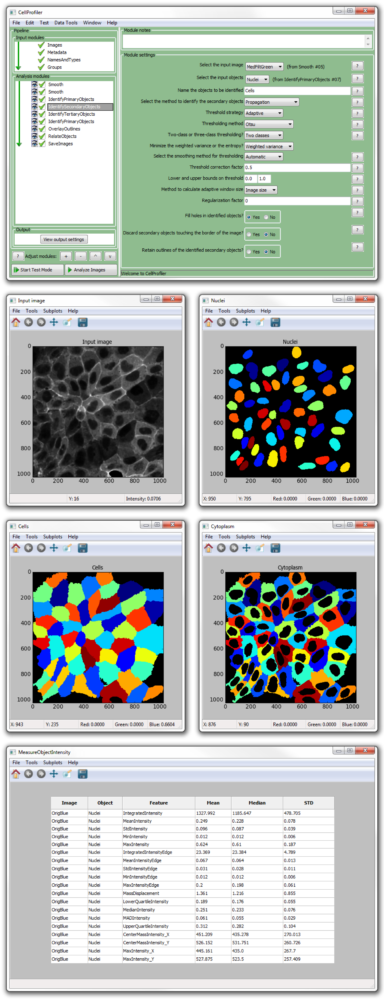
Online resources for biological image analysis, including software These resources are more comprehensive review articles that cover the latest developments in the broader world of biological image analysis, including analysis for three-dimensional image stacks, time-lapse images, analysis of whole organisms, and imaging modalities like brightfield microscopy, differential-interference-contrast imaging, electron microscopy, and biomedical imagery (MRI and PET scans of humans or model organisms, for example). We do not attempt a comprehensive review of biological image analysis but instead point the reader to excellent resources in the field (see Box 1). This is only an introductory taste of how image analysis works, exemplified by one particular application area. Carpenter, and Michael Yaffe, unpublished data). We and our collaborators have used this assay to screen chemical compounds and genes (using RNA interference) in human cells and Drosophila melanogaster cells to identify regulators of DNA-damage-response pathways (Scott Floyd, Michael Pacold, Thouis R. The foci are labeled by an antibody that recognizes the phosphorylated form of a protein that responds to DNA damage. The goal in this assay is to identify samples where cells show an unusually strong or unusually weak response to DNA damage by counting the number of DNA-damage-induced foci per cell. Throughout this tutorial, we will use the example of a cell-based fluorescence microscopy assay for DNA-damage regulators ( Figure 1). Colors of nuclei, cells, and foci are arbitrary. Schematic data shown, based on image courtesy of Scott Floyd, Michael Pacold, and Michael Yaffe. Nuclei are identified by thresholding, then used as seeds to identify cell edges. Overall image analysis workflow for a typical experiment.įirst, variations in illumination and staining are corrected.
Computers have also been able to distinguish the subtle differences between localization patterns that seem identical to a human investigator. For example, researchers typically cannot distinguish cells in the G1 phase of the cell cycle from those in G2 by looking at images of DNA-stained cells, but automated algorithms can do so by quantifying the fluorescence intensity of the DNA in each nucleus. Researchers have also identified situations where automated image analysis can “see” phenotypes invisible to humans. In the case of hundreds of phenotype-relevant genes or chemicals discovered in a single screen, the quantitative measurement of multiple cellular phenotypes enables those samples to be sorted into distinct subtypes for further analysis and characterization, as has been done recently for mitotic-spindle defects and defects in cytokinesis. There are other substantial scientific benefits, however: automated image analysis can yield objective and quantitative measurements, thereby enabling the capture of subtle differences among samples as well as statistical analysis and systems-biology research on the data. Often the goal of automated image analysis is simply to replicate a human's observations with less labor. As a second example, counting dozens of DNA-damage-induced foci in each of hundreds of cells in each of tens of thousands of images would simply be impossible by eye yet automated image analysis enabled such a screen to identify regulators of DNA-damage responses (Scott Floyd, Michael Pacold, Thouis R. This enables experiments on an entirely different scale than before for example, an automatically analyzed microscopy screen of the human genome by RNA interference (more than 300,000 images) recently revealed many classes of mitosis-essential genes in multiple phenotypic categories. The most obvious advantage of automated image analysis is speed, especially now that automated microscopes can capture images faster than a human can examine them. Yet there are many cases where scoring visual phenotypes with a computer is highly attractive. Annotation of such complex and varied phenotypes is beyond the capabilities of current computer software.
Presentation of images in cellprofiler analyst manual#
One impressive example is a genome-wide RNA-interference screen for dozens of phenotypes, where extensive manual annotation of more than 40,000 movies of early embryogenesis in Caenorhabditis elegans uncovered the detailed involvement of hundreds of genes in development. Although examining tens of thousands of samples by eye is tedious, biologists are often highly motivated to invest the effort in order to discover samples of interest or annotate large sets of chemically or genetically perturbed samples. Microscopy is one of the foundational tools of biology, and researchers have for centuries relied on their own visual systems to interpret what they see.


 0 kommentar(er)
0 kommentar(er)
